June 16th 2022
DeepSARS: Sequencing and testing at the same time
A new scientific platform called DeepSARS, developed by Prof Sai Reddy, BRCCH Vice Director and Professor of Systems and Synthetic Immunology at ETH Zurich, could support viral testing and tracking for future pandemics: While current diagnostic testing and genomic surveillance methods are being done separately, DeepSARS is able to perform both tasks simultaneously. This allows for earlier detection of emerging variants and profiling mutations at the population level. In this conversation, Prof Reddy joins journalist Irène Dietschi to discuss the consortium’s exciting new findings.
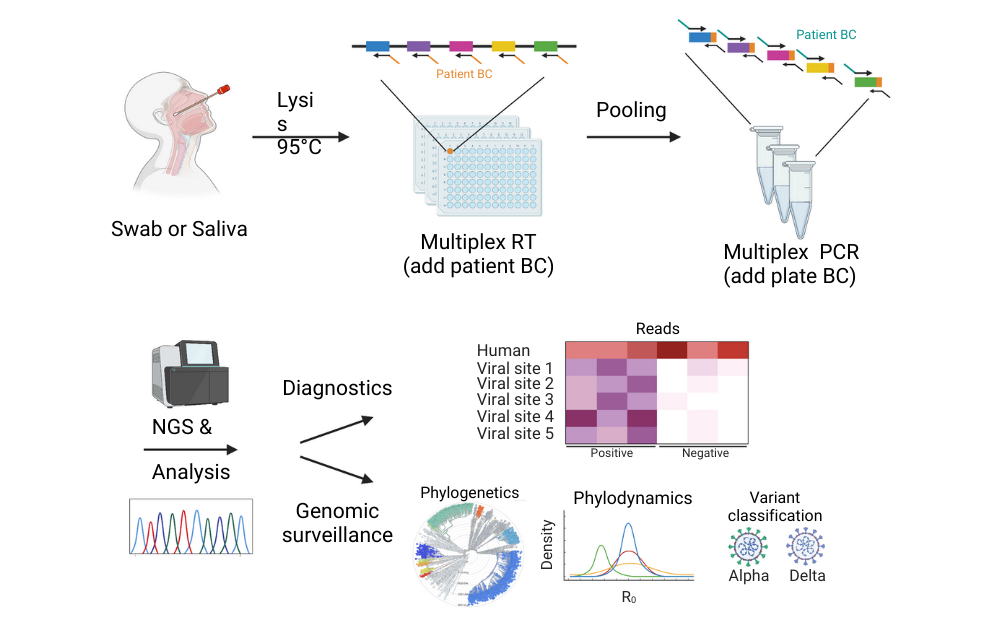
Graphical abstract: DeepSARS uses molecular barcodes (BCs) and multiplexed targeted deep sequencing (NGS) to enable simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2. Image courtesy of Yermanos et al. 2022
The ongoing COVID-19 pandemic remains a major global health concern with novel variants of SARS-CoV-2, such as alpha, beta, gamma, delta and omicron, that are continuously emerging and resulting in new waves of infection. Potentially exacerbating the situation, many of the masking and social distancing rules are relaxed around the world.
The latest variant of SARS-CoV-2, omicron, with over 50 mutations is able to infect at a greater capacity than previous strains and often possesses substantial immune evasion, leading to many breakthrough infections in vaccinated or previously infected individuals.
Genomic surveillance of SARS-CoV-2 has been a vital component in the monitoring of the pandemic, providing valuable information for guiding public health decisions. However, despite the fact that the SARS-CoV-2 genomes from infected patients have been sequenced at unprecedented levels, they still represent a small fraction of the total number of global infections. And countries vary widely in how they prioritize viral genome sequencing. What makes it even more problematic is that in the countries with low sequencing rates, they also tend to have high infection rates and are therefore at an especially high risk for new mutations to evolve. They are hotspots for new variants.
Key region: the receptor binding domain
That is where DeepSARS comes into play. "A number of researchers have previously embraced the idea that sequencing could be used not just for genomic surveillance, but also for diagnostic testing", says immunology professor Sai Reddy, Vice Director of the BRCCH. Prof Reddy, who is the principal investigator of the Laboratory for Systems and Synthetic Immunology at ETH Zurich, has actually pursued this idea to a practical level. In a paper published recently in BMC Genomics, he, his team and collaborators have established a scientific concept which has proven that sequencing and testing can be done simultaneously. Their system is called DeepSARS.
"One of the main challenges of the project was to determine which sites in the viral genome would yield maximum diagnostic and genomic information while maintaining sufficient coverage for each site", explains Prof Reddy. One region which is most associated with new variants is the spike protein of SARS-CoV-2, and within the spike protein it is particularly the so-called receptor binding domain – the site where the virus attaches itself to the host receptor ACE2 – where mutations most likely occur.
This receptor binding domain also happens to be the target site of neutralizing antibodies, generated from either vaccination or previous infection. Mutations which emerge from this region can influence variants’ attachment to host cells, potentially making them more transmissible and are allowing them to escape from neutralizing antibodies. "Therefore, when we are developing sequencing tests, it is very important to get information about the receptor binding domain", says Prof Reddy. "It was a central part for the design of DeepSARS."
Experimental protocol and ‘targeted deep sequencing’
Once this was established, Sai Reddy and colleagues proceeded in a stepwise manner to develop the DeepSARS system. "First we elaborated an experimental protocol, which is in fact very similar to the protocol in which all PCR tests are performed", says Prof Reddy. "The main difference is that we added little personalized markers to each sample of the PCR, so that when you perform a genomic sequencing experiment you know which patient it came from." This procedure is called molecular barcoding.
The second step the researchers undertook was to identify regions for what they call "targeted deep sequencing". Prof Reddy: "Targeted deep sequencing means reducing the viral genome from 30’000 RNA bases to about 20 percent of that number - 6000 bases. And it means identifying those 20 percent of sites which give us information that will likely track the evolution of the virus, as well as identify emerging variants." To achieve that, Reddy and colleagues performed an extensive computational analysis (i.e., computational phylogenetics). In this way they identified target candidates of the viral genome and then implemented them in the experimental protocol.
Proof of concept on patients’ sample
Next, they validated their data on synthetic RNA templates of SARS-CoV-2, based on genomic sequences recovered early in the pandemic. "That allowed us to do very precise experiments to detect the amount of viral material, which was as low as 10 copies of the virus per sample", Prof Reddy explains.
The final step was testing DeepSARS on samples from patients. "We had samples from 30 patients, and we were able to show, based on nasal swabs or saliva, that the DeepSARS testing assay was very close to performing at the same level of diagnostic detection as a traditional PCR test", says Prof Reddy. And: "It was able to provide enough information about genomic surveillance to classify viral evolution, and whether there were any variants or not." In short: DeepSARS works. With the proof of concept, its science and technology may be regarded as well established.
A solid foundation for future pandemics
The exciting question is, could DeepSARS be applied at a larger scale? Could it even be deployed in the current COVID-19 pandemic? "DeepSARS certainly has a lot of potential", says Prof Reddy. "But right now, while we’re still in the COVID-19 pandemic, it wouldn’t make much sense to replace the current infrastructure for diagnostics and genomic surveillance with a new system." As Prof Reddy explains, that would implicate numerous changes in the logistics and regulations of the on-going pandemic, which, among other aspects, would be far too costly and require extensive regulatory approval.
Yet Sai Reddy is collaborating with public health experts in Switzerland as well as with clinicians at University Hospital Basel and other partners, discussing future applications, including clinical testing of DeepSARS. The goal is to examine the system at a larger scale. "We have shown that DeepSARS can be rapidly adapted for identification of emerging variants and for profiling mutational changes at a small scale but for a pandemic, this requires population level implementation", says Prof Reddy. "Practically speaking, DeepSARS could be of immense benefit in for future pandemics or possibly as SARS-CoV-2 transitions to an endemic stage."
Background
DeepSARS was developed by a BRCCH-supported consortium where bioengineers, immunologists, computational biologists and clinical scientists work together and as part of a larger BRCCH research programme, the Fast Track Call for COVID-19 research (FTC). The programme aims to enable research that will help mitigate medical and public health challenges and contribute tangible solutions to reduce global disease burden due to COVID-19.
This specific project aims for rapid transfer of these state-of-the art diagnostic methods across Switzerland and many other countries around the world, leading the way for innovative population level surveillance approaches of future variants and other disease outbreaks.
Interview: Irène Dietschi
Research article:
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08403-0
Yermanos A, Hong KL, Agrafiotis A, Han J, Nadeau S, Valenzuela C, Azizoglu A, Ehling R, Gao B, Spahr M, Neumeier D, Chang CH, Dounas A, Petrillo E, Nissen I, Burcklen E, Feldkamp M, Beisel C, Oxenius A, Savic M, Stadler T, Rudolf F & Reddy ST. 2022. "DeepSARS: simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2." BMC Genomics. DOI:v10.1186/s12864-022-08403-0
MORE about the BRCCH-supported COVID-19 research by Prof Sai Reddy and consortium.